Scalable Biologically-Aware Skeleton Generation for Connectomic Volumes
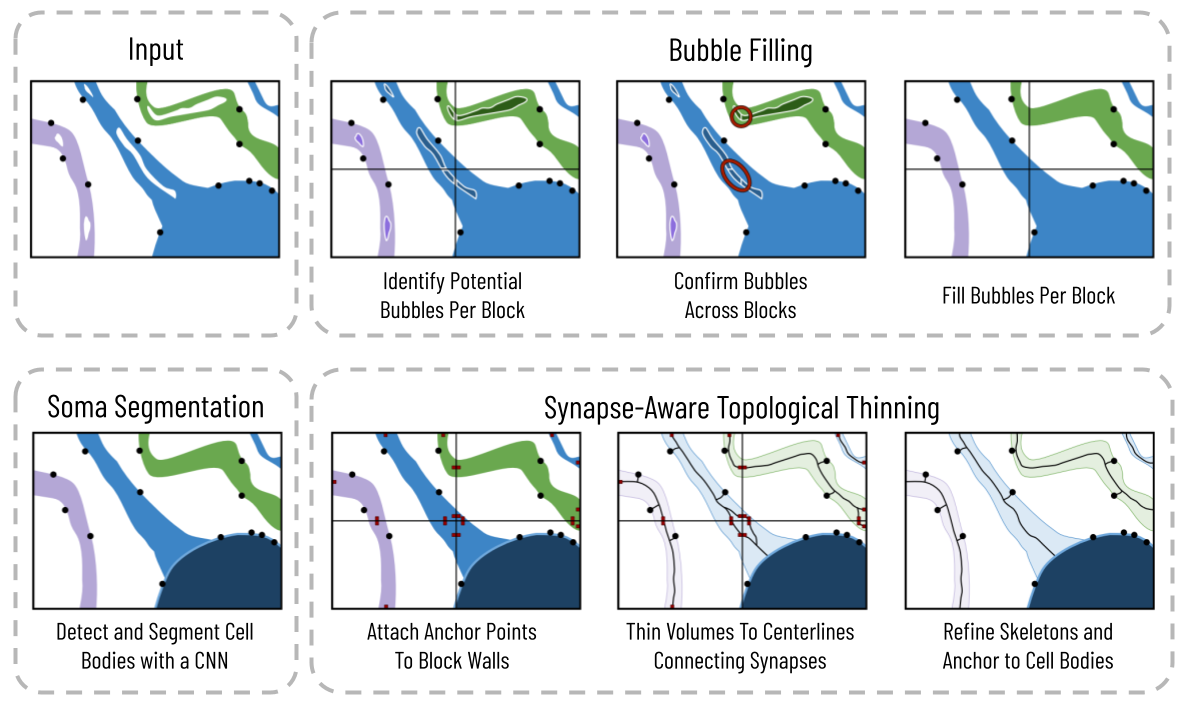
As connectomic datasets exceed hundreds of terabytes in size, accurate and efficient skeleton generation of the label volumes has evolved into a critical component of the computation pipeline used for analysis, evaluation, visualization, and error correction. We propose a novel topological thinning strategy that uses biological-constraints to produce accurate centerlines from segmented neuronal volumeshile still maintaining biologically relevant properties. Current methods are either agnostic to the underlying biology, have non-linear running times as a function of the number of input voxels, or both. First, we eliminate from the input segmentation biologically-infeasible bubbles, pockets of voxels incorrectly labeled within a neuron, to improve segmentation accuracy, allow for more accurate centerlines, and increase processing speed. Next, a Convolutional Neural Network (CNN) detects cell bodies from the input segmentation, allowing us to anchor our skeletons to the somata. Lastly, a synapse-aware topological thinning approach produces expressive skeletons for each neuron with a nearly one-to-one correspondence between endpoints and synapses. We simultaneously estimate geometric properties of neurite width and geodesic distance between synapse and cell body, improving accuracy by 47.5% and 62.8% over baseline methods. We separate the skeletonization process into a series of computation steps, leveraging data-parallel strategies to increase throughput significantly. We demonstrate our results on over 1250 neurons and neuron fragments from three different species, processing over one million voxels per second per CPU with linear scalability.