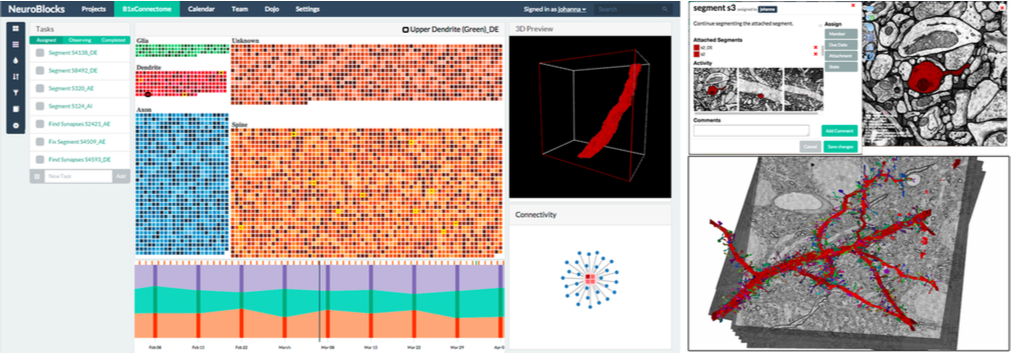
In the field of connectomics, neuroscientists acquire electron microscopy volumes at nanometer resolution in order to reconstruct a detailed wiring diagram of the neurons in the brain. The resulting image volumes, which often are hundreds of terabytes in size, need to be segmented to identify cell boundaries, synapses, and important cell organelles. However, the segmentation process of a single volume is very complex, time-intensive, and usually performed using a diverse set of tools and many users. To tackle the associated challenges, this paper presents NeuroBlocks, which is a novel visualization system for tracking the state, progress, and evolution of very large volumetric segmentation data in neuroscience. NeuroBlocks is a multi-user web-based application that seamlessly integrates the diverse set of tools that neuroscientists currently use for manual and semi-automatic segmentation, proofreading, visualization, and analysis. NeuroBlocks is the first system that integrates this heterogeneous tool set, providing crucial support for the management, provenance, accountability, and auditing of large-scale segmentations. We describe the design of NeuroBlocks, starting with an analysis of the domain-specific tasks, their inherent challenges, and our subsequent task abstraction and visual representation. We demonstrate the utility of our design based on two case studies that focus on different user roles and their respective requirements for performing and tracking the progress of segmentation and proofreading in a large real-world connectomics project.
Our IEEE SciVis 2015 paper is available here or at our webpage.